David Blei’s Machine Learning Algorithms May One Day Be a Tool in Doctors’ Kits
By analyzing massive genetic data sets, algorithms can help identify disease-carrying mutations; by analyzing health records, they can help predict an individual’s survival.
In artificial intelligence, often the challenge is finding enough data to train the system to interpret information. In the case of understanding disease from reading the genomes of populations—and thus expediting efforts to tailor health care to an individual's DNA—the problem is finding algorithms that can uncover meaningful patterns within the massive genetic data sets that are already available.
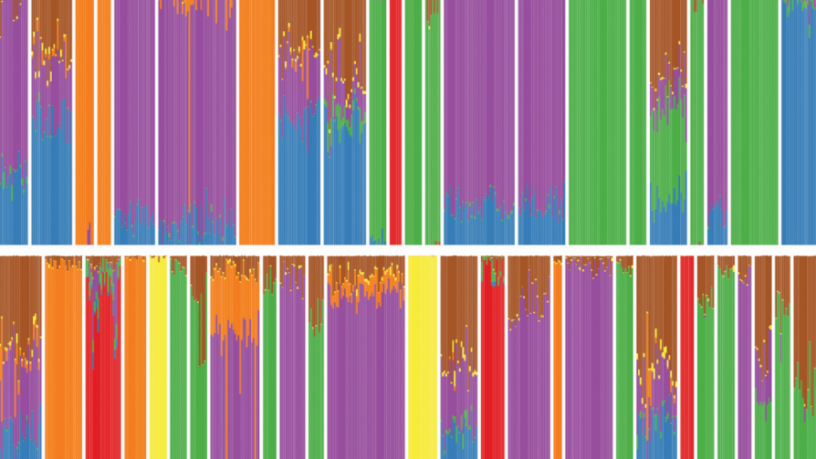
Using TeraStructure, David Blei can estimate population structures. Each vertical slice represents one person’s mix of ancestral populations. (Image courtesy of Wai Hao)
The relationship between genes and traits, which must be understood before personalized medicine can advance, is confounded by population structure. Ancestral populations all reproduced together and then migrated across the earth. A team of Columbia and Princeton researchers led by Columbia Computer Science Professor David Blei and Princeton statistician John Storey has developed a new machine learning algorithm that can scan the enormous quantities of genetic data randomly dispersed across populations. On simulated data sets of 10,000 people, the algorithm, dubbed TeraStructure, could estimate population structure twice as fast as current state-of-the-art algorithms. TeraStructure could analyze the genomes of one million individuals, orders of magnitude beyond modern software capabilities, the researchers said, and could potentially characterize the structure of world-scale human populations.

David Blei (Photo by Jeffrey Schifman)
The researchers' algorithm builds on the widely used and adapted Structure algorithm, a Bayesian model. The Structure algorithm cycles through an entire data set, genome by genome, one million variants at a time, before updating its model both to characterize ancestral populations and to estimate their proportion in each individual. Repeated passes through the data set refine the model.
By contrast, TeraStructure, an application of another Blei advance called stochastic variational inference, updates the model as it goes. It samples one genetic variant at one location and compares it to all variants in the data set at the same location across the data set, producing a working estimate of population structure. “You don’t have to painstakingly go through all the points each time to update your model,” Blei said.
Blei’s team is employing TeraStructure in other health-related projects, in collaboration with researchers at Columbia’s College of Physicians and Surgeons. Chief among them is an effort to expand survival analysis to large-scale data by parsing the thousands of electronic health records on file at NewYork-Presbyterian Hospital. The researchers are actively building new tools for monitoring patients and predicting the course of their disease based on outcome data made available by the analysis.