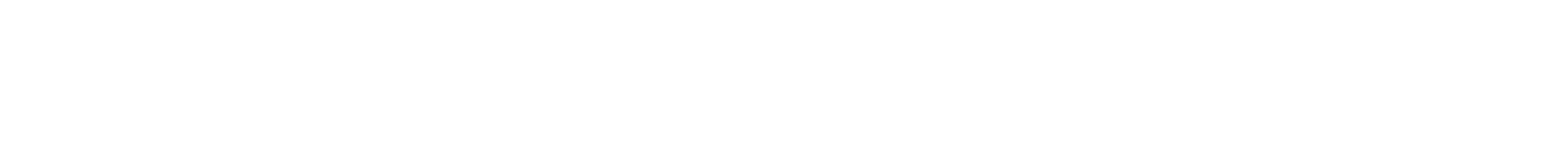Faculty Q&A: Joining Forces
Engineers and clinicians target new fronts in the fight to eradicate cancer
OCT scanner for imaging breast specimens.
Engineering is integral to modern clinical practice, designing the technology that increasingly empowers cutting-edge medicine. Perhaps nowhere is that more evident than in the rapidly evolving area of cancer research, as evidenced by the strong partnerships between Columbia Engineering, Columbia’s Herbert Irving Comprehensive Cancer Center (HICCC), and the Columbia University Irving Medical Center (CUIMC).
As those ties broaden and deepen, we spoke to a few of our researchers putting innovative diagnostic and treatment tools in the hands of clinicians.
Gordana Vunjak-Novakovic engineers novel “organs-on-a-chip” platforms. A University Professor and the Mikati Foundation Professor of Biomedical Engineering and Medicine, Vunjak-Novakovic collaborates with medical faculty including Andrea Califano, Clyde and Helen Wu Professor of Chemical and Systems Biology, to unlock the mechanics of metastatic progression.
Christine Hendon devises next-generation imaging tools and algorithms to extract their key data. Hendon, an associate professor of electrical engineering, works with Hanina Hibshoosh, professor of pathology and cell biology, and Richard Ha, associate professor of radiology, on solutions for earlier detection.
Tal Danino programs microbes for novel behaviors. An associate professor of biomedical engineering, Danino joined with Nick Arpaia, assistant professor of microbiology and immunology, to develop promising strategies for drug delivery via engineered bacteria.
Elham Azizi, assistant professor of biomedical engineering, creates machine learning methods to mine cell data. With Brent Stockwell, professor of biological sciences, and Jellert Gaublomme, assistant professor of biological sciences, she’s investigating the role of intercellular interactions in treatments. With Ran Reshef, associate professor of medicine, she’s looking at complications from stem cell transplants.
In a wide-ranging roundtable with Columbia Engineering magazine, they discussed modulating the immune system, overcoming the limitations of classic models, and the revolutionary potential of artificial intelligence (AI).

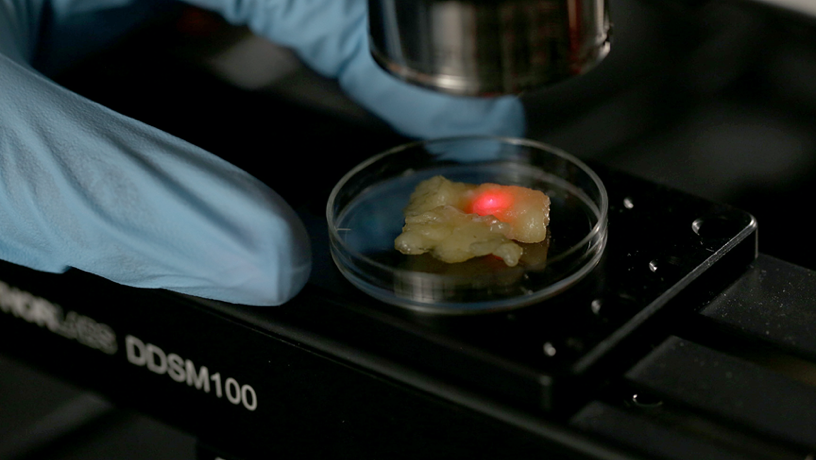
Single-cell Map of Breast Tumor-Immune Microenvioronment.
How well do we understand the tumor microenvironment? How are engineering and computational approaches positioned to address that challenge?

Tal Danino.
Tal Danino: The tumor microenvironment is extremely complex. From the numerous cell types to matrices, stroma, and synapses, there is a delicate interplay of multiple factors. As engineers, we focus on identifying these components both in isolation and in context. We create distilled, controlled ex vivo models to further understand the role of each factor. While we can engineer a system to partially reflect this complex space, computational approaches allow for the modeling of many more factors and provide insight into the dynamics of the more complete system. Specifically, our group studies the interaction between bacteria and tumors using a three-dimensional tumor spheroid system to better mimic the architecture of the tumor microenvironment.

Gordana Vunjak-Novakovic.
Gordana Vunjak-Novakovic: Despite advances, many patients still don’t respond to therapy or relapse after treatment, with metastatic progression being the major cause of these poor outcomes. Tumor metastasis is particularly difficult to study, both in a clinic and in cancer cell cultures and animal models. Over the last decade, bioengineering has begun developing human models of cancer: organoids and tissue-engineered tumor models. While these models are a major advance over cell culture and animal models, they still fail to recapitulate the critically important interactions of tumor cells with the host tissues that can regulate tumor development or serve as metastatic sites, and the immune and vascular cells. To be predictive and useful to the field, a bioengineered model should emulate the complexity of cancer progression, from primary tumor growth to cancer cell intravasation into the bloodstream and extravasation into distal organ sites, and should incorporate vascular and immune cells. Moreover, the ability to model clinically observed cancer progression and treatment in an individualized, patient-specific fashion is critical for understanding cancer heterogeneity and the underlying differences between responders and non-responders to cancer therapy. Our laboratory is now developing these models in collaboration with Andrea Califano, Hanina Hibshoosh, Kevin Kalinsky, and Peter Sims.
One roadblock to early detection is simply the cumbersomeness of processing biopsies. AI and imaging potentially have a huge role to play in streamlining this process. Still, something as basic as differentiating “regions of interest” from “regions of non-interest” is a major hurdle to automation. How does your team leverage different expertise to tackle that?

Christine Hendon.
Christine Hendon: Our team is a collaboration between electrical engineering, radiology, and pathology. The development of our AI algorithms and advances in our optical imaging system are guided by the needs identified by Drs. Ha and Hibshoosh. Dr. Ha routinely performs biopsies and reviews medical images for breast diagnosis. His feedback has been instrumental in interpreting optical coherence tomography (OCT) images of breast specimens. During early meetings, he pointed out features within our OCT images of specimens that looked similar to features within ultrasound images. This helped in developing initial machine learning algorithms. Dr. Hibshoosh has given us feedback on our experimental design and also our optical system parameters, as we strive to have both ultra-high-speed and ultra-high-resolution OCT imaging. After conducting imaging experiments, we review our histology slides with him to help us identify interest versus non-interest. The challenge is that there is so much diversity within and between patients. Therefore, large datasets are needed to develop an algorithm that performs well and can be generalized to a testing set. Our team, including investigators and many engineering graduate students, meets weekly.
Hanina Hibshoosh: Key to our collaboration is the integration and tolerance of diverse perspectives. This spans from defining a clinical problem, engineering a workable solution and proving its utility. The path however is frequently not linear and orderly as described and benefits from patience and at time suspension of first impressions of applicability and utility. No part can do it alone and the whole is greater than the sum of its parts. Assembling the right team to address a problem is key.
What AI concepts from other realms do you think show the most promise for translating to cancer therapies? How are you working to adapt them?

Elham Azizi.
Elham Azizi: There are multiple concepts in AI that can significantly help us in understanding and dissecting the complexity of cancer. My work has been mainly focused on developing probabilistic models for unsupervised learning of cell states in the tumor microenvironment. This modeling approach provides the advantages of interpretability and evaluating confidence in predictions and estimations. We have adapted the generative process in these models to mimic biological processes such as gene regulation and thus have been able to resolve technical artifacts in data and integrate multiple types of data to learn the underlying circuitry of cell types. Deep neural networks in AI are also powerful for developing methods that are scalable to the growing size of datasets. Recently we have started building novel deep generative models to leverage both interpretability and scalability in characterizing low-dimensional manifolds representing diverse cell states in the tumor microenvironment.
As the technologies mature, what does the partnership between AI and medical professionals look like?
Richard Ha: AI technology will help in clinical decision making by giving us more information that can help personalize treatment plans for each patient. It’s going to augment rather than replace the role of a medical professionals.
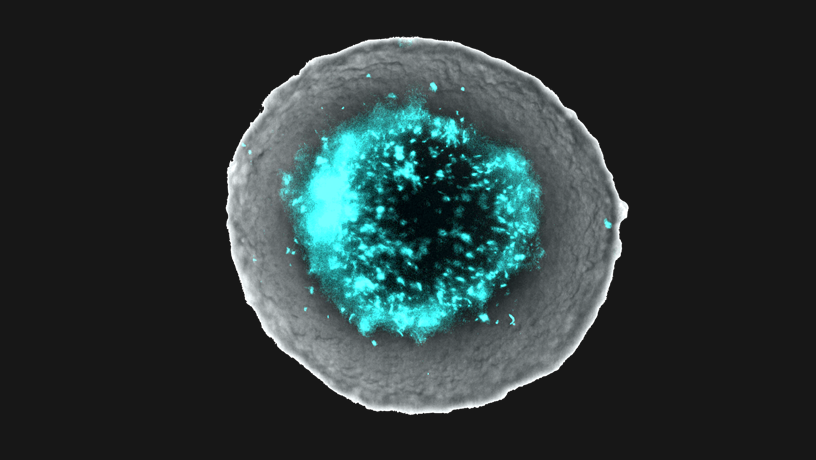
Co-culture of engineered bacteria within tumor spheroids.
More and more, feedback from clinical observations influences what happens in the laboratory. How have you seen that play out in your work?
CH: My laboratory started with developing automated algorithms and new high-resolution systems for imaging the heart. Columbia provides a great collaborative environment, where I was able to meet Dr. Hibshoosh, and later, Dr. Ha. They provide feedback on desired imaging speed and image resolution to ensure that any system and algorithm that we develop fits within the clinical workflow.
TD: As engineers, we aim to innovate where there is a clinical need. One example is using our bacteria approach to circumvent toxicities seen in checkpoint therapies clinically. While antibodies targeting immune checkpoints are therapeutically effective in regressing some cancers, they can result in severe side effects. Motivated by this limitation of an otherwise promising therapy, we sought to recombinantly produce therapies against these proven targets and locally deliver the drugs, thereby reducing toxicities.
GVN: The best starting point for conducting a research project is when our clinician colleagues contact us about addressing a specific clinical problem. From the onset, we start working together and continue so throughout the project, to get inputs on our designs and their validation against clinical data.
One such technology is the organs-on-a-chip platform that contains miniature human tissues grown to establish the necessary levels of maturity and function and connected to each other by vascular perfusion. Organs-on-a-chip is a fast-track application of tissue engineering as they can immediately advance many areas of medicine without long and expensive clinical trials. By encompassing innovation in human stem cell technology, organs-on-a-chip offer to emulate human pathophysiology in vitro—including patient-specific studies of cancer—and overcome some of the limitations of the cell and animal models.
Importantly, the goal is to establish the minimally functional units that can recapitulate certain aspects of human physiology in a controlled and straightforward manner.
How are engineering approaches helping uncover the balance between personalizing therapeutic interventions and investigating underlying mechanisms that cut across different pathologies?
GVN: Both aspects are incredibly important in the context of cancer and other systemic pathologies. Our knowledge about the regulators of tumor progression and response to treatment, as well as about any commonalities between different pathologies, is still evolving. I believe that the patient-specific tissue models of cancer that are now being developed and validated against clinical data will allow quantitative studies of personalized therapeutic interventions by using patient-specific models of cancer progression and treatment and to investigate the underlying mechanisms for clinically observed responses.
TD: One of the primary reasons we are excited by the prospect of bacterial cancer therapies is their potential of being tumor type agnostic. Bacteria preferentially colonize the hypoxic and necrotic microenvironment, present in virtually all solid tumors. Therefore, bacteria can successfully colonize tumors in multiple tissue types, regardless of antigen-expression or tumor genetics. To this end, we are able to deliver a variety of therapeutics using our bacterial delivery system including toxins, as well as immunotherapeutic nanobodies and cytokines
What are the implications for technologies that can modulate the immune system, even beyond cancer research?

Nick Arpaia.
Nick Arpaia: There are numerous tissues within the body that are now know to harbor an endogenous microbiome. Aside from the gut, recent studies suggest that diverse communities of microbes are naturally present in the skin, other organs such as the pancreas and lung, as well as within tumors. Using probiotic bacteria as a chassis for drug delivery, we hope to apply synthetic biology principles to target the bacteria to specific sites of disease and deliver immunomodulatory agents for the treatment of allergic and autoimmune disease, and metabolic disorders as well as infectious diseases.
What are some recent breakthroughs to come out of your collaborations?
GVN: Working with Drs. Califano, Sims, Hibshoosh, and Kalinsky, and a team of talented young investigators, our laboratory has designed a “cancer patient on a chip” model of invasive human breast carcinoma. The platform allows physiological integration of the tumor with its cognate metastatic sites (lung, liver, bone with bone marrow) via vascular perfusion that contains circulating cancer and immune cells. The tumor compartment can be established from surgical specimens to preserve the phenotype and heterogeneity of cancer cells. At the same time, the target metastatic sites and vasculature can be bioengineered, starting from blood-derived, patient-matched iPS cells. In studies of drugs, the liver compartment also serves as the site of first-pass drug metabolism.
We seek to recapitulate the key aspects of human pathophysiology in quantitative studies of tumor progression and response to treatment. Our focus is on breast cancer metastasis, particularly hormone positive and triple-negative invasive ductal carcinomas, the most common aggressive cancer in women, which lacks effective therapeutic modalities.
CH: We recently had a manuscript published in the Journal of Academic Radiology that showed that using a convolutional neural network, we could classify ultra-high-resolution OCT images of normal breast tissue from cancer with 94 percent accuracy. We are looking forward to continuing to add to our database so that we can increase the diversity of our dataset. We would like to have a large range of breast cancer types and of normal tissue, acquired from many patients, to ensure that any algorithm we develop can be disseminated to the wider community.
HH: We are poised to formally prove that our imaging technique combined with AI interpretation can transform our capacity to assess tissues at the gross stage of clinical analysis. This has a large range of applications at every stage of the diagnostic process: biopsy, intraoperative excision, gross assessment, tumor banking and is applicable across multiple organs and diseases.