That’s Life: Engineers Make and Exploit Living Molecules
Molecular engineering—at the intersection of the life sciences, engineering, and computer science—is about making new molecules. Researchers in this hybrid field seek to understand the fundamental processes of life; at the same time, they exploit that knowledge to modify and synthesize biological materials for solutions to real-world problems.
Columbia molecular engineers, particularly genomics researchers, are paving the way for achievements in radical new approaches to medicine, such as gene editing and gene therapy, that are still in their infancy.
Recent years have also seen a resurgence of interest in immunotherapy, which marshals the body’s own immune system to fight disease, especially cancer. Developed in the late nineteenth century by William Coley, the “father of immunotherapy,” the approach fell out of use in favor of today’s familiar cancer treatments of surgery, radiation, and chemotherapy. Today, molecular engineering researchers at Columbia are making exciting advances toward the creation of immunotherapeutic agents, including synthetic vaccines against cancer and deadly viruses.
Molecular engineering also holds great promise in nonmedical areas, such as digital information storage, water purification, and agriculture.
Bacteria: Engineering’s Newest Research and Clinical Collaborators
“Being able to manipulate life.” That, in five words, is what drew Tal Danino to synthetic biology. And not just for the intellectual adventure, but as a way to engineer solutions to real-world problems.
Danino, assistant professor of biomedical engineering at Columbia Engineering and director of the Synthetic Biological Systems Laboratory, wants to put bacteria to work detecting and treating diseases such as cancer. The heart of his technology lies in programming the DNA in bacteria to make diagnostic and therapeutic molecules from inside the tumor.
Immune cells cannot enter the tumor’s center, called the “necrotic core.” Bacteria like the environment in the core: not only are they safe, but they also can feed on nutrients provided by dead cancer cells.
“Not only can we make certain small molecules,” says Danino, “but we can also make a lot of other things. We can make proteins and enzymes that cause holes in cancer cell membranes—that’s what bacteria do; they kill cells. We also can make small RNA molecules, to silence genes that are expressed in cancer.”
Danino has engineered a probiotic to detect liver cancer. When the proliferating bacteria sense the tumor environment, depending on which genetic circuit used, they produce a molecule that either changes the color of the urine or causes it to luminesce. Now his goal is to make probiotics that can get into tumors, sense the environment, and release a therapeutic drug in response.
In late July, Nature published a paper coauthored by Danino on recent work using attenuated, or less virulent, bacteria to treat liver tumors. He and his colleagues used the bacteria to deliver three different genetic circuits into liver tumors: one to produce a molecule that attacks tumor cell membranes, one to produce a molecule that causes cells to self-destruct, and one to produce a protein to stimulate the immune system.

In mouse tests, used alone the bacteria reduced tumor growth slightly, but combining the bacteria with a common chemotherapy drug, 5-fluorouracil, caused the tumor to shrink much more than when only the drug was used.
Danino also expresses his creativity through bio-art projects, recently in collaboration with Brazilian artist Vik Muniz. Some artworks consist of actual bacteria; others use a photo. In perhaps the most striking work, the bacteria are alive and growing while exhibited. For Danino, science and art are not mutually exclusive.
“It is all about exploration; about being comfortable with uncertainty and seeing where it leads you,” he says.
Tal Danino is a member of the Data Science Institute.

Marshaling Millions of Web Collaborators
 Yaniv Erlich may have the largest research group of any scientist. His collaborators are the more than 20,000 people who have so far posted their genomes on the crowdsourced online genetic database DNA.Land. Site users upload genomes that they have obtained from one of three consumer genetic-testing companies—23andMe, FamilyTreeDNA, or Ancestry.com—providing Erlich and his colleagues with a wealth of data.
Yaniv Erlich may have the largest research group of any scientist. His collaborators are the more than 20,000 people who have so far posted their genomes on the crowdsourced online genetic database DNA.Land. Site users upload genomes that they have obtained from one of three consumer genetic-testing companies—23andMe, FamilyTreeDNA, or Ancestry.com—providing Erlich and his colleagues with a wealth of data.
rlich, assistant professor of computer science at the Engineering School, with a joint appointment at the New York Genome Center (NYGC), started the site with fellow NYGC member Joseph Pickrell, adjunct assistant professor of biological sciences at Columbia. Erlich’s genomics research, including his cutting-edge gene-sequencing algorithms, motivated him to harness the resources of the online world as a way to obtain as many human genomes as possible, both efficiently and cost-effectively. Additionally, he liked the idea of giving people a way to be involved with his work.
No stranger to untraditional data-collection methods, in 2013 Erlich used genealogical data harvested from social media sites to create a family tree of 13 million people, going back to the 15th century.
Though some genetic-testing companies provide researchers with information, that information is an aggregate, not connected to individual genomes. DNA.Land is unique among online genetic databases in that users can link their genomes to their social media profiles, providing further information on how genes affect human health. For example, a user may complain on Facebook about being sleep deprived during the workweek. Users also fill out surveys on their ancestry and health.
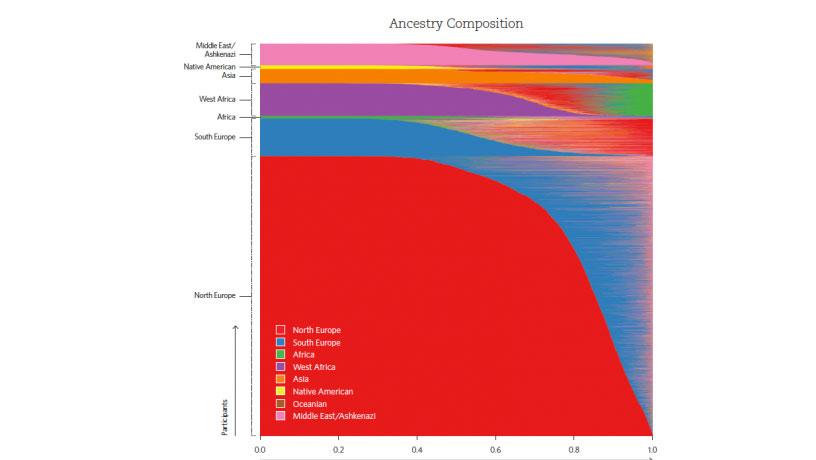
Because each genetic-testing company tests for different genetic markers, DNA.Land uses a technique called “imputation” to infer genetic variants not included in a specific test. The technique is based on the phenomenon that certain markers are frequently inherited together. “In return for their genomes,” says Erlich, “DNA.Land offers users more than just a sense of virtue for having contributed to science.” Users receive a “contribution badge,” showing points accumulated for answering questions.They will be updated with new findings about their genome. They also receive information about their ancestry and relatedness to other users. (Erlich discovered a fourth cousin.) Of course, they may learn disquieting information, such as what the site refers to as a “non-paternity event” in the family.
Erlich and his colleagues are sensitive to privacy issues. DNA.Land informs users that there is always a low risk of a confidentiality breach. Making the same contribution they ask of users, Erlich and Pickrell posted their own genomes on DNA.Land.
When asked if he would have posted his genome on DNA.Land had he had a mutation linked with a serious late-onset disease, Erlich does not hesitate before responding. “Privacy is important but the whole point of posting genomes is to find solutions for serious diseases. If you have a serious mutation, would you not want the best science to understand it?”
Yaniv Erlich is a member of the Data Science Institute.