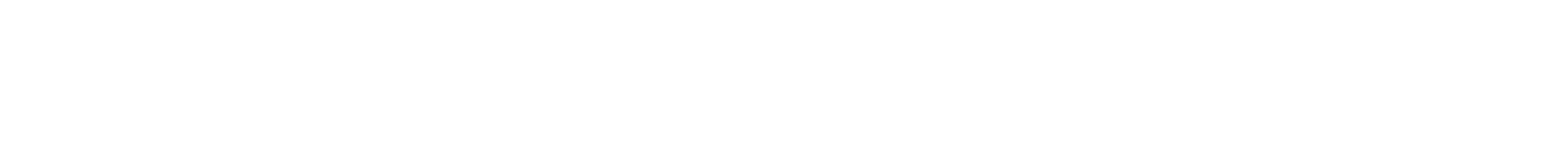Is DNA the Next Hard Drive?
A new coding strategy maximizes DNA data-storage capacity for the first time.
As Yaniv Erlich cruised along on the New York City subway one afternoon, trying to get cell reception, a novel idea began to take shape. A computational biologist and computer science professor, Erlich routinely works at the intersection between artificial and biological code. Now, he began to wonder, could algorithms designed for streaming video on smartphones be the key to unlocking a new approach for storing data in biological processes?
We humans generate a staggering amount of data—some experts estimate we’ll hit 44 trillion gigabytes by 2020—with the potential to soon outstrip the total capacity of hard drives and magnetic tape combined.
For almost a decade, researchers in search of a solution have been tantalized by the possibilities inherent to nature’s age-old technique for information storage—DNA. But no one has been able to devise a method capable of exploiting the full extent of the double helix’s tremendous capacity.
Erlich’s insight was to employ the phone’s erasure-correcting algorithm, called fountain codes, to open up nearly all DNA’s potential by squeezing more information into its four nucleotide bases. His team demonstrated that this technology is also extremely reliable.

Erlich (left) with collaborator Dina Zielinski, a colleague at the New York Genome Center.
DNA Encoding Sequence.
DNA is an ideal storage medium because it’s ultracompact and can last hundreds of thousands of years if kept in a cool, dry place, as demonstrated by the recent recovery of DNA from the bones of a 430,000-year-old human ancestor found in a cave in Spain. As an added bonus, outside the encoding and retrieval process the medium also requires no energy to hold the information.
“DNA won’t degrade over time, and, unlike cassette tapes and CDs, it won’t become obsolete,” said Erlich, who is also a member of Columbia’s Data Science Institute and the New York Genome Center (NYGC).
Along with Dina Zielinski, a colleague at NYGC, Erlich chose six files to encode, or write, into DNA: a full computer operating system; an 1895 French film, Arrival of a Train at La Ciotat; a $50 Amazon gift card; a computer virus; a Pioneer plaque; and a 1948 study by information theorist Claude Shannon.
They compressed the files into a master file and then split the data into short strings of binary code made up of ones and zeros. Using the fountain codes, they randomly packaged the strings into so-called droplets and mapped the ones and zeros in each droplet to the four nucleotide bases in DNA: A, G, C, and T. The algorithm deleted letter combinations known to create errors and added a barcode to each droplet to help reassemble the files later.
In all, they generated a digital list of 72,000 DNA strands, each 200 bases long, and sent it in a text file to a San Francisco DNA-synthesis startup, Twist Bioscience, that specializes in turning digital data into biological data. Two weeks later, they received a vial holding a speck of DNA molecules.
To retrieve their files, they used modern sequencing technology to read the DNA strands, followed by software to translate the genetic code back into binary code. They recovered their files with zero errors, the study reports.
DNA Decoding Sequence.
They also demonstrated that a virtually unlimited number of copies of the files could be created with their coding technique by multiplying their DNA sample through polymerase chain reaction (PCR) and that those copies, and even copies of their copies, and so on, could be recovered error-free.
But the real excitement came when the researchers showed that their coding strategy packs 215 petabytes of data on a single gram of DNA—100 times more than methods published by pioneering researchers George Church at Harvard and Nick Goldman and Ewan Birney at the European Bioinformatics Institute. “We believe this is the highest-density large-scale data-storage architecture ever created,” said Erlich.
The capacity of DNA data storage is theoretically limited to two binary digits for each nucleotide, but the biological constraints of DNA itself, and the need to include redundant information to reassemble and read the fragments later, reduces its capacity to 1.8 binary digits per nucleotide base.
The fountain codes were what made this advance possible. A technique Erlich remembered from graduate school, these codes streamlined the reading and writing process so efficiently that Erlich and Zielinski could pack an average of 1.6 bits into each nucleotide base. That’s at least 60 percent more data than previously published methods, and close to the 1.8-bit limit.
Now that Erlich’s team has proof the technique works, the next step will be to develop a secure version of the technology. Funded by a DARPA grant, Erlich is joining forces with three Columbia engineers—cryptologists Tal Malkin and Eran Tromer, along with Itsik Pe’er, who focuses on computational genetics—to create a biological key that will ensure access for legitimate users, while preventing adversaries from cloning the DNA.
“The idea is to create a mathematical framework to define a very explicit set of properties by which we could ensure security of the system,” Erlich said. “Then we’ll go back to the chemical side to develop these properties.”
This spring, he’s also organizing an invitation-only symposium in New York that will bring together some two dozen researchers from around the world to discuss new applications for DNA storage.